Department of Mycobacterium Reference and Research
The Department of Mycobacterium Reference and Research (DMRR) is composed of two divisions, namely bacteriology and molecular epidemiology. These two divisions are primarily conducting studies on mycobacteriology, including pathogen-environment-patient interrelations. Currently, seven organic researchers are carrying out studies on their own areas of expertise. These areas of expertise include pathogenicity, morphology (ultrastructure analysis), genetics, and diagnostics for clinical management of mycobacterial diseases.
DMRR is also recognized as a substantial national reference laboratory for mycobacteria in Japan, and works as a supra-national reference laboratory in the WHO Western Pacific Region.
Bacteriology Division
The bacteriology division mainly conducts mycobacteriological research. We focus on bacteriological aspects of mycobacteriosis; the development of new diagnostics, the analyses of drug-resistant Mycobacterium tuberculosis, microscopic morphological studies by electron microscope, and quality assurance of mycobacteriological techniques (external quality assessment of smear microscopy with artificial sputum specimens, drug susceptibility testing, etc.).
DMRR serves as the national reference laboratory for mycobacteria. Our division performs genetic analysis of drug-resistant M.tuberculosis, species identification of M.tuberculosis and MOTT (Mycobacterium Other Than Tuberculosis), etc.
Colonies of Mycobacteria on Ogawa’s medium

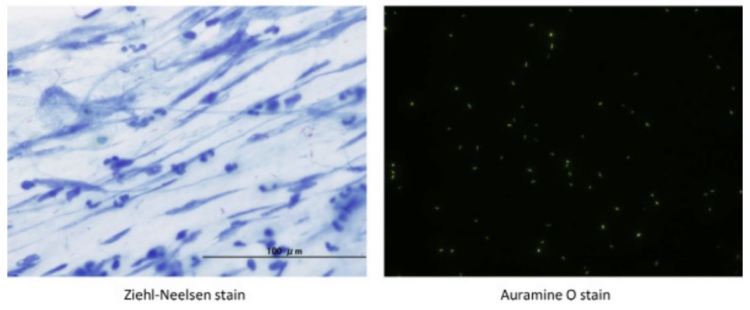
Sputum microscopy of Mycobacterium tuberculosis
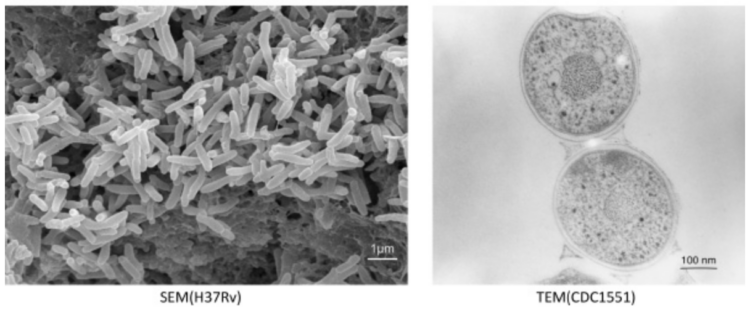
Electron microscopy of Mycobacterium tuberculosis
Molecular Epidemiology Division
The Molecular Epidemiology division studies the development and improvement of genotyping methods for mycobacteria. The techniques are utilized in the molecular epidemiology of Mycobacterim tuberculosis, studies for the pathogenicity analysis of nontuberculosis mycobacteria (in collaboration with Fukujuji Hospital).
Molecular epidemiological study of tuberculosis
M. tuberculosis strains from different patients have polymorphisms on genomic DNA, and we can distinguish different strains of M. tuberculosis by using genotyping methods, including VNTR, spoligotyping, IS6110-RFLP, and whole genome sequence comparison.
TB genotyping results, when combined with epidemiological data, can help to understand the epidemiology of TB infections in a given setting. Below are some applications of molecular epidemiological study:
- To confirm epidemiological linkage of outbreak suspected cases;
- To identify unknown or unsuspected transmission settings, such as internet cafes, language schools and hostels, instead of traditional settings such as homes and offices;
- To identify the emergence and spreading of specific strains such as MDR/XDR-TB;
- To detect laboratory cross-contamination event; and
- To understand the genetic diversity of tuberculosis strains circulating in Japan and their virulence.
Quality assurance of VNTR
Quality assurance of genotyping is important for molecular epidemiological surveying. We have been conducting proficiency testing on VNTR in Japan. We also hold training courses on VNTR annually.
Species identification of Mycobacteria
Though more than 150 nontuberculosis mycobacterial (NTM) species were reported, identifiable species in clinical laboratories are quite limited. We have been investigating the prevalence of NTM species in Japan by accepting unidentifiable NTM species from clinical laboratories. We use 16S rRNA, rpoB, hsp65 and ITS sequences for species identification of NTM.
